Identifying the Issue
- In silico technique to replace or assist and reduce the cumbersome experimental techniques in protein engineering.
- In silico validation using mutations with known functional implications in homologs.
Objective of the Research
- To use in silico techniques as a replacement for experimental techniques for protein engineering.
- To validate the findings using experimentally verified mutations with known functional implications from homologous proteins.
Who should read this?
It is helpful for all the experimentalists who are interested in protein engineering and understanding the role of single nucleotide polymorphisms from genomic studies after high throughput analysis.
Solution
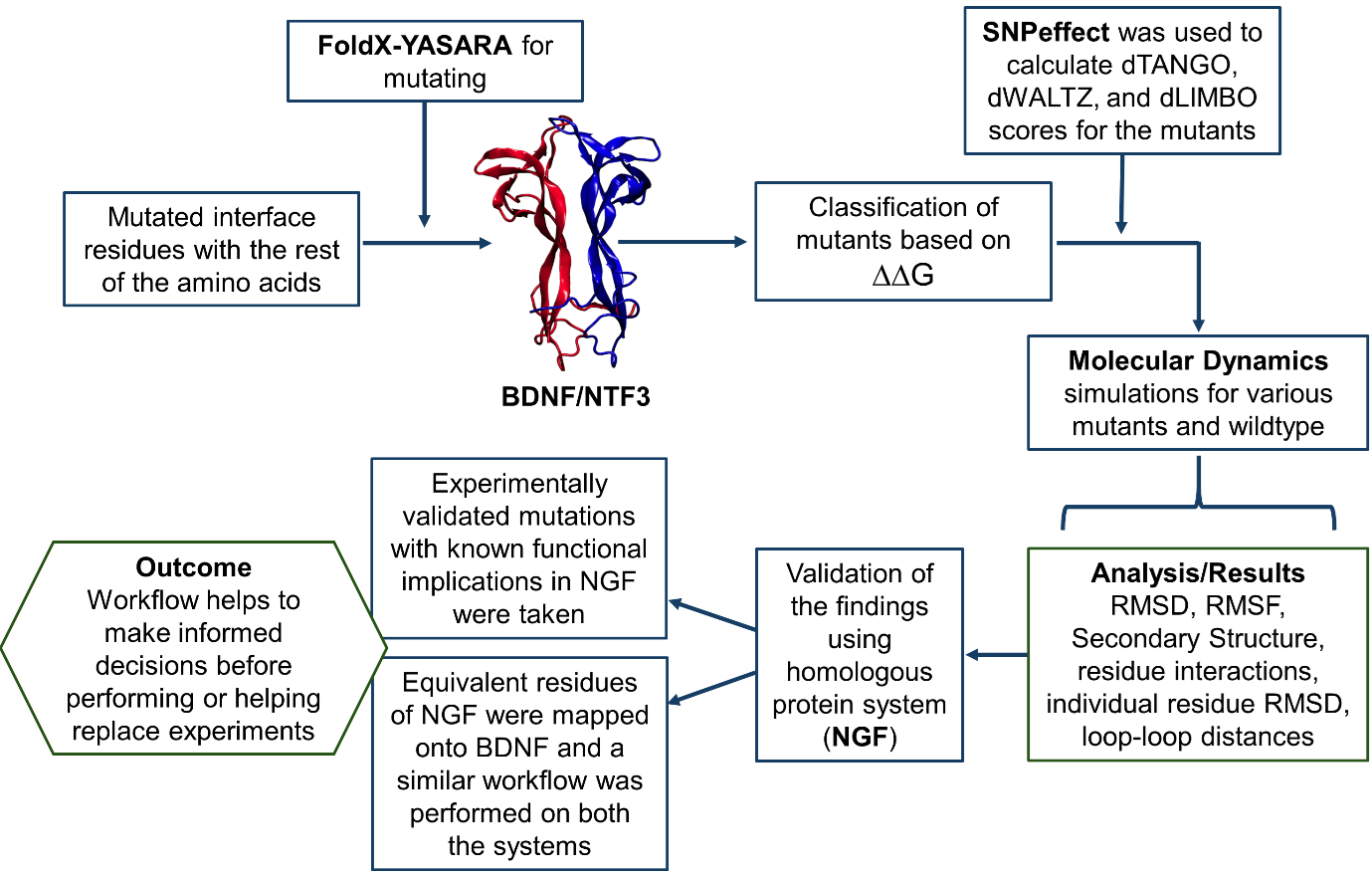
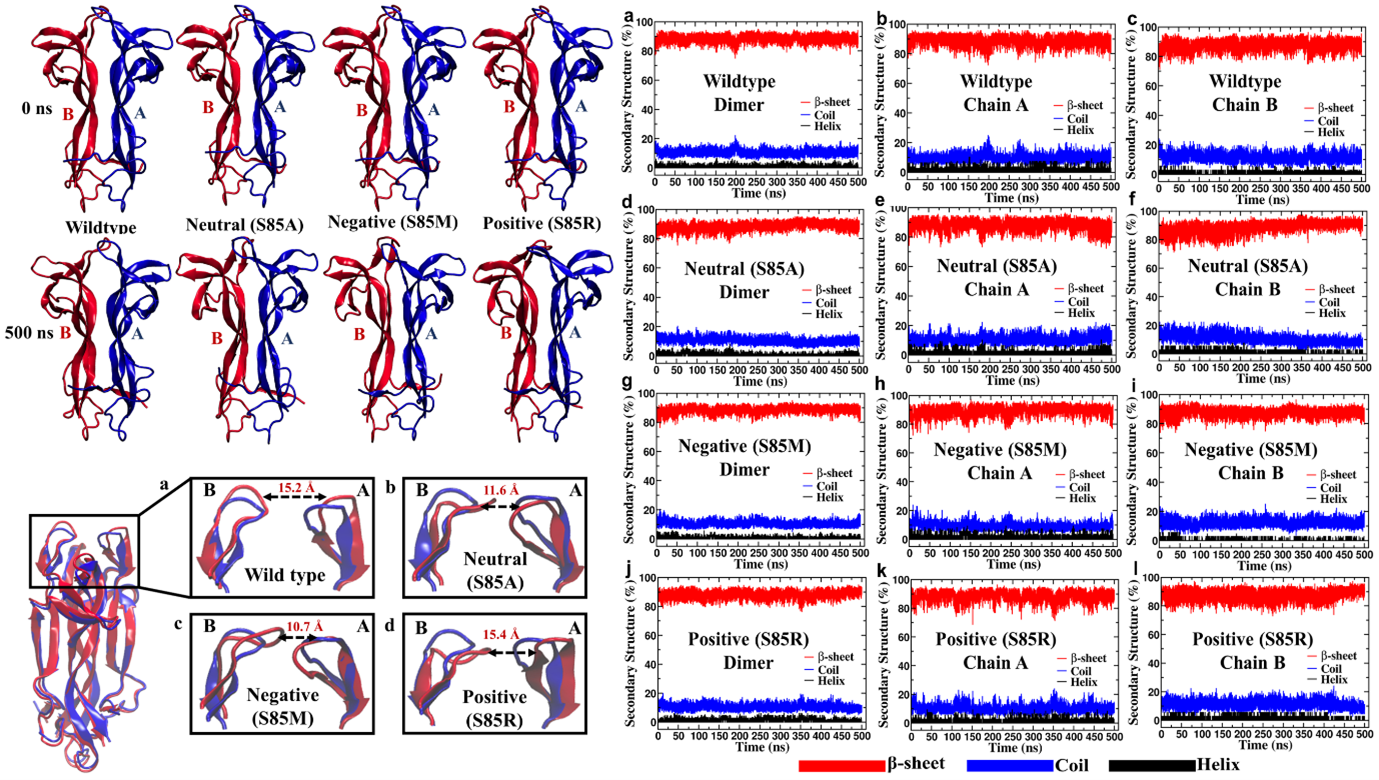
Protein engineering by directed evolution is time-consuming. Hence, in silico techniques using software for ∆∆G calculation, and predicting propensity for aggregation, amyloid formation, and chaperone binding are employed to design proteins. We employed these techniques to engineer BDNF-NTF3 interactions. The effect of different mutations was screened for their favourable, unfavourable, and lack of effect in the BDNF NTF3 system. The unfavourable mutants showed fewer hydrophobic interactions and higher hydrogen bonds compared to the wild-type or other mutants with similar solvent-accessible surface area, suggesting solvent-mediated disruption of hydrogen-bonded interactions.
The predictions in the BDNF system were validated using mutations with known functional effects in the NGF system where similar results were obtained. The results suggest that mutations with known effects in homologous proteins could help validation, and in silico-directed evolution, experiments could be a viable alternative to or for arriving at informed decisions or as a supplement to the experimental technique used for protein engineering.
Key Features and Benefits
- High throughput data generation for informed decisions to aid protein engineering.
- Validation from the experimentally available data using in silico
- The method can help to understand the effect of single nucleotide polymorphisms from the genomic data.
Impact
- This workflow helps to make informed decisions before performing or helping replace experiments.
- This workflow will help to reduce the cumbersome experiments required in protein engineering.
- The method can help to understand the effect of single nucleotide polymorphisms from the genomic data.
- Validation from the experimentally available data using in silico methods/techniques.
Team
- V.M. Datta Darshan, Department of Biosciences, Sri Sathya Sai Institute of Higher Learning
- Venketesh Sivaramakrishnan, Department of Biosciences, Sri Sathya Sai Institute of Higher Learning
- Subbarao Kanchi, Department of Physics, Sri Sathya Sai Institute of Higher Learning
- Natarajan Arumugam, Department of Chemistry, College of Science, King Saud University
- Abdulrahman I. Almansour, Department of Chemistry, College of Science, King Saud University
Title of paper: “In silico energetic and molecular dynamic simulations studies demonstrate potential effect of the point mutations with implications for protein engineering in BDNF”
Read Paper Here: https://doi.org/10.1016/j.ijbiomac.2024.132247